Estimate transfer function models by Inverse Filtering.
Source:R/armax.inverse.fit.R
armax.inverse.fit.Rdarmax.inverse.fit(
DATA,
order = hydromad.getOption("order"),
delay = hydromad.getOption("delay"),
fit.method = hydromad.getOption("inverse.fit.method"),
normalise = TRUE,
init.U = TRUE,
pars = NULL,
use.Qm = TRUE,
fft.inverse.sim = FALSE,
rises.only = FALSE,
...,
max.iterations = hydromad.getOption("inverse.iterations"),
rel.tolerance = hydromad.getOption("inverse.rel.tolerance"),
par.epsilon = hydromad.getOption("inverse.par.epsilon"),
init.attempt = 0,
trace = hydromad.getOption("trace")
)Arguments
- DATA
a
ts-like object with named columns:- list("Q")
observed output time series.
- list("P")
observed input time series (optional).
- order
the transfer function order. See
armax.- delay
delay (lag time / dead time) in number of time steps. If missing, this will be estimated from the cross correlation function.
- fit.method
Placeholder
- normalise
Placeholder
- init.U
Placeholder
- pars
Placeholder
- use.Qm
Placeholder
- fft.inverse.sim
Placeholder
- rises.only
Placeholder
- ...
Placeholder
- max.iterations
Placeholder
- rel.tolerance
Placeholder
- par.epsilon
Placeholder
- init.attempt
Placeholder
- trace
Placeholder
Value
a tf object, which is a list with components
- coefficients
the fitted parameter values.
- fitted.values
the fitted values.
- residuals
the residuals.
- delay
the (possibly fitted) delay time.
Details
In normal usage, one would not call these functions directly, but rather
specify the routing fitting method for a hydromad model using
that function's rfit argument. E.g. to specify fitting an
expuh routing model by inverse filtering one could write
hydromad(..., routing = "expuh", rfit = "inverse")
or
hydromad(..., routing = "expuh", rfit = list("inverse", order =
c(2,1))).
References
...
See also
armax.inverse.sim,
expuh, armax.sriv.fit
Examples
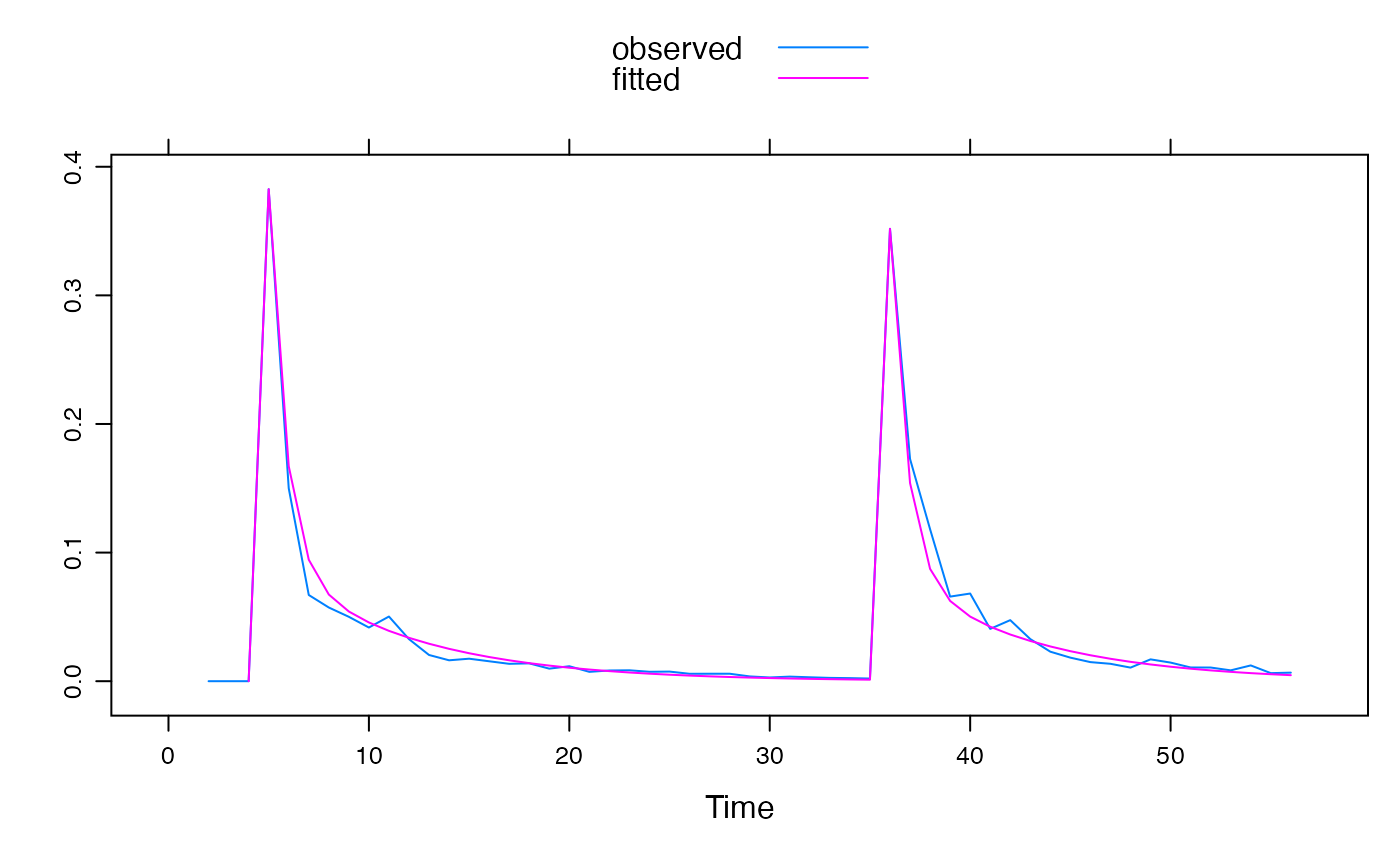
U <- ts(c(0, 0, 0, 1, rep(0, 30), 1, rep(0, 20)))
Y <- expuh.sim(lag(U, -1), tau_s = 10, tau_q = 2, v_s = 0.5, v_3 = 0.1)
set.seed(0)
Yh <- Y * rnorm(Y, mean = 1, sd = 0.2)
fit1 <- armax.inverse.fit(ts.union(P = U, Q = Yh),
order = c(2, 2), warmup = 0
)
#> Warning: did not converge after 12 iterations
fit1
#>
#> Unit Hydrograph / Linear Transfer Function
#>
#> Call:
#> armax.inverse.fit(DATA = ts.union(P = U, Q = Yh), order = c(2,
#> 2), warmup = 0)
#>
#> Order: (n=2, m=2) Delay: 1
#> ARMAX Parameters:
#> a_1 a_2 b_0 b_1 b_2
#> 1.148917 -0.246043 0.348228 -0.247966 -0.003136
#> Exponential component parameters:
#> tau_s tau_q v_s v_q v_3
#> 6.85247 0.79598 0.62715 0.38560 -0.01275
#> TF Structure: S + Q + inst. (three in parallel)
#> Poles:0.2847, 0.8642
#>
xyplot(ts.union(observed = Yh, fitted = fitted(fit1)),
superpose = TRUE
)