Run many simulations by sampling within parameter ranges.
Arguments
- object
a
hydromadobject (produced by thehydromad()function) that is not fully specified (i.e. so some parameter values are given as ranges).- nsim
number of parameter samples to run.
- seed
optional random seed, for repeatability.
- ...
further arguments to
FUN.- sampletype
sampling method; see
parameterSets.- FUN
optional function to apply to each simulated model. Typical examples would be
objFunVal,summaryorpredict.- objective
an objective function (or statistic function); this is just an argument to be passed on to
FUN, which in this case defaults toobjFunValto calculate the statistic value from each model. It is treated as a special argument because it is cached before the simulation run for efficiency.- bind
to bind the result from
FUNas one or more columns onto the matrix of parameter values, and return as a data frame.
Value
a list of results, where each element is named by its parameter set.
The result also has an attribute "psets" which gives the parameter
values used in each simulation (as a data frame).
If bind = TRUE, a data frame.
Details
none yet.
See also
Examples
data(Canning)
mod0 <- hydromad(Canning[1:500, ], sma = "cwi")
sim0 <- simulate(mod0, nsim = 5, sampletype = "latin")
coef(sim0)
#> tw f scale l p t_ref
#> tw=50, f=2, l=0, p=1, t_ref=20 50 2 1.177428e-05 0 1 20
#> tw=0, f=4, l=0, p=1, t_ref=20 0 4 2.159142e-04 0 1 20
#> tw=100, f=6, l=0, p=1, t_ref=20 100 6 8.978107e-06 0 1 20
#> tw=25, f=0, l=0, p=1, t_ref=20 25 0 4.535322e-05 0 1 20
#> tw=75, f=8, l=0, p=1, t_ref=20 75 8 8.949261e-06 0 1 20
summary(sim0)
#> rel.bias r.squared r.sq.sqrt r.sq.log
#> tw=50, f=2, l=0, p=1, t_ref=20 -6.792001e-17 -0.2964138 -0.3549607 -0.4302310
#> tw=0, f=4, l=0, p=1, t_ref=20 1.720709e-17 -1.6340737 -0.7640808 -0.4968276
#> tw=100, f=6, l=0, p=1, t_ref=20 8.077254e-17 -0.4916033 -0.4320486 -0.4671808
#> tw=25, f=0, l=0, p=1, t_ref=20 5.923279e-18 -0.2566781 -0.2921281 -0.3663058
#> tw=75, f=8, l=0, p=1, t_ref=20 -2.450985e-17 -0.4956284 -0.4333150 -0.4676961
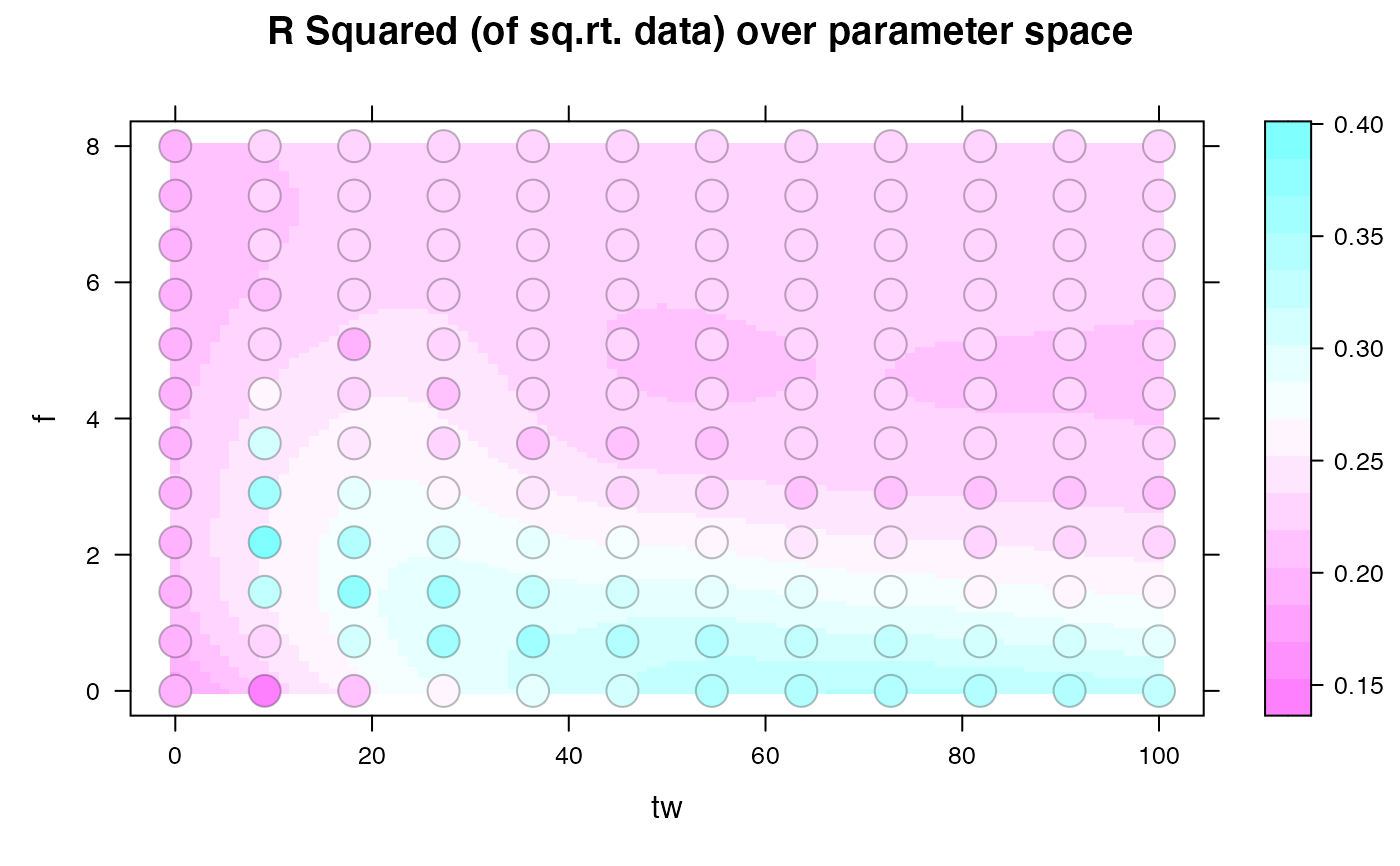
## plot the objective function surface over two parameters
mod1 <- update(mod0, routing = "armax", rfit = list("ls", order = c(2, 1)))
sim1 <- simulate(mod1, 144,
sampletype = "all", FUN = objFunVal,
objective = ~ nseStat(Q, X, trans = sqrt)
)
levelplot(result ~ tw + f, sim1,
cex = 2,
panel = panel.levelplot.points,
main = "R Squared (of sq.rt. data) over parameter space"
) +
latticeExtra::layer(panel.2dsmoother(...), under = TRUE)
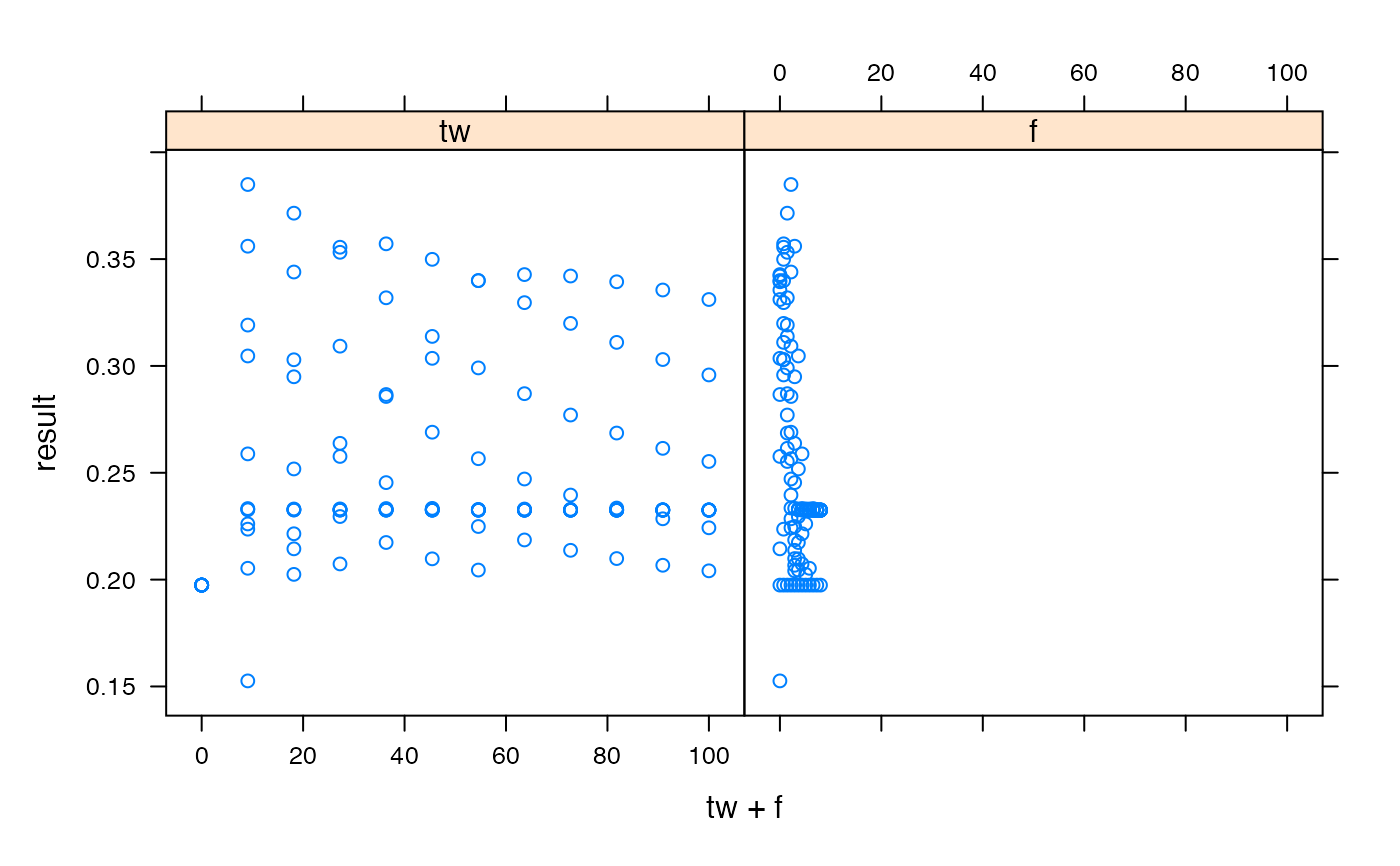
 ## dotty plots (list any number of parameters in formula)
xyplot(result ~ tw + f, sim1, outer = TRUE)
## dotty plots (list any number of parameters in formula)
xyplot(result ~ tw + f, sim1, outer = TRUE)