Plot methods...
# S3 method for hydromad
plot(x, y, ...)
# S3 method for hydromad
xyplot(
x,
data = NULL,
...,
scales = list(),
feasible.bounds = FALSE,
col.bounds = "grey80",
border = "grey60",
alpha.bounds = 1,
all = FALSE,
superpose = TRUE,
with.P = FALSE,
type = "l",
type.P = c("h", if ("g" %in% type) "g"),
layout = c(1, NA)
)
# S3 method for hydromad.runlist
xyplot(
x,
data = NULL,
...,
scales = list(),
all = FALSE,
superpose = FALSE,
with.P = FALSE,
type = "l",
type.P = c("h", if ("g" %in% type) "g"),
layout = c(1, NA)
)
# S3 method for hydromad
qqmath(
x,
data = NULL,
...,
all = FALSE,
type = "l",
auto.key = list(lines = TRUE, points = FALSE),
f.value = ppoints(100),
tails.n = 100
)
# S3 method for hydromad
tsdiag(object, gof.lag, ...)Arguments
- x
an object of class
hydromad.- y
Placeholder for plot.hydromad
- ...
further arguments passed on to
xyplot.zooorqqmath.- data
ignored.
- scales
Placeholder
- feasible.bounds
if
TRUE, then ensemble simulation bounds are extracted and plotted. This only works if a feasible set has been specified usingdefineFeasibleSetor theupdatemethod. Note that the meaning depends on what value ofglue.quantileswas specified to those methods: it might be the overall simulation bounds, or some GLUE-like quantile values.- col.bounds, border, alpha.bounds
graphical parameters of the ensemble simulation bounds if
feasible.bounds = TRUE.- all
passed to
fitted()andobserved().- superpose
to overlay observed and modelled time series in one panel.
- with.P
to include the input rainfall series in the plot.
- type
Placeholder
- type.P
plot type for rainfall, passed to
panel.xyplot.- layout
Placeholder
- auto.key
Placeholder
- f.value, tails.n
arguments to
panel.qqmath.- object, gof.lag
passed to the
arimamethod oftsdiag.
Value
the trellis functions return a trellis object.
See also
hydromad.object, xyplot,
xyplot.ts, xyplot.list
Examples
data(Canning)
cannCal <- window(Canning, start = "1978-01-01", end = "1982-12-31")
mod <-
hydromad(cannCal,
sma = "cwi", tw = 162, f = 2, l = 300,
t_ref = 0, scale = 0.000284,
routing = "expuh", tau_s = 4.3, delay = 1, warmup = 200
)
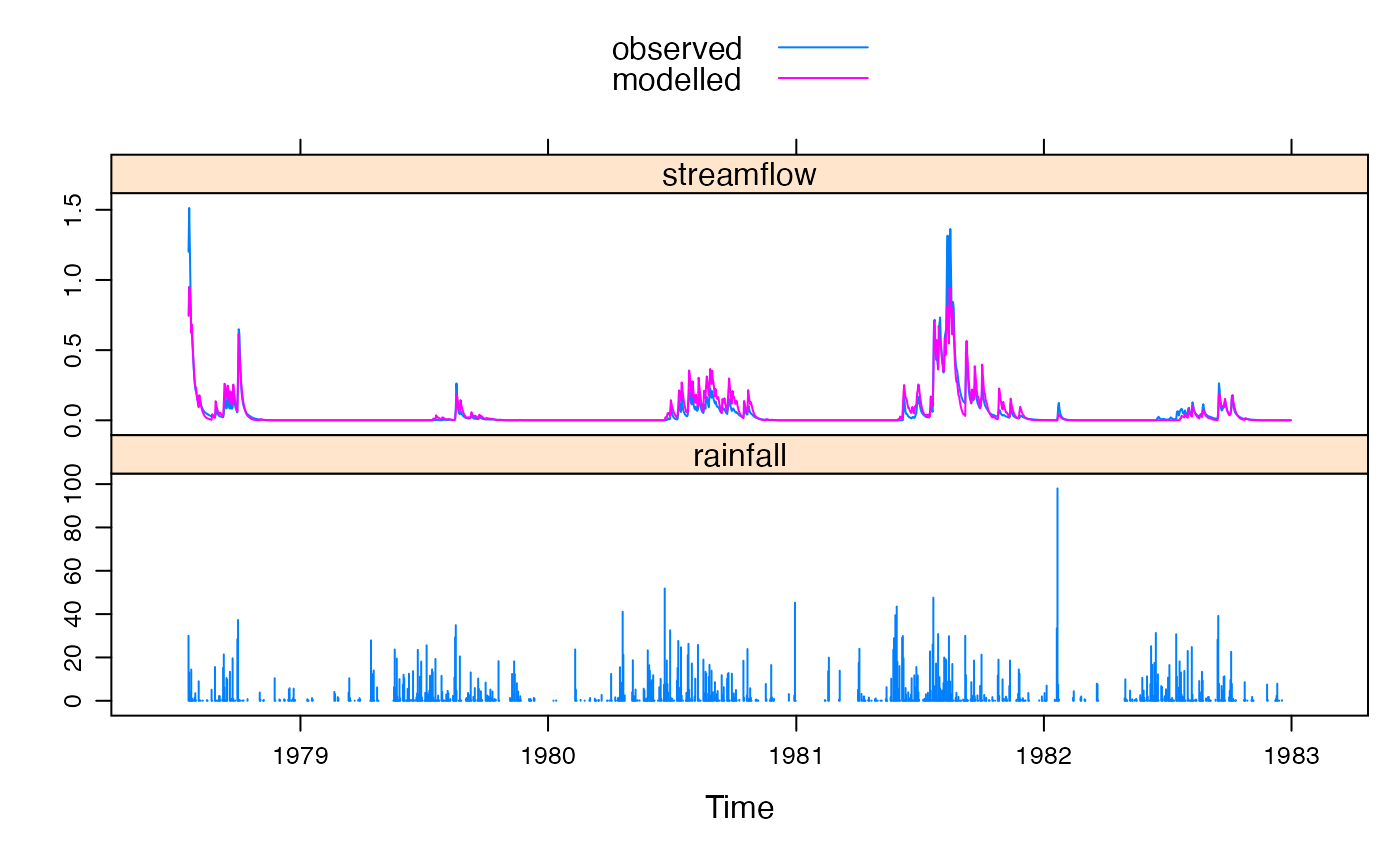
xyplot(mod, with.P = TRUE)
 c(
streamflow = xyplot(mod),
residuals = xyplot(residuals(mod, type = "h")),
layout = c(1, 2), y.same = TRUE
)
c(
streamflow = xyplot(mod),
residuals = xyplot(residuals(mod, type = "h")),
layout = c(1, 2), y.same = TRUE
)
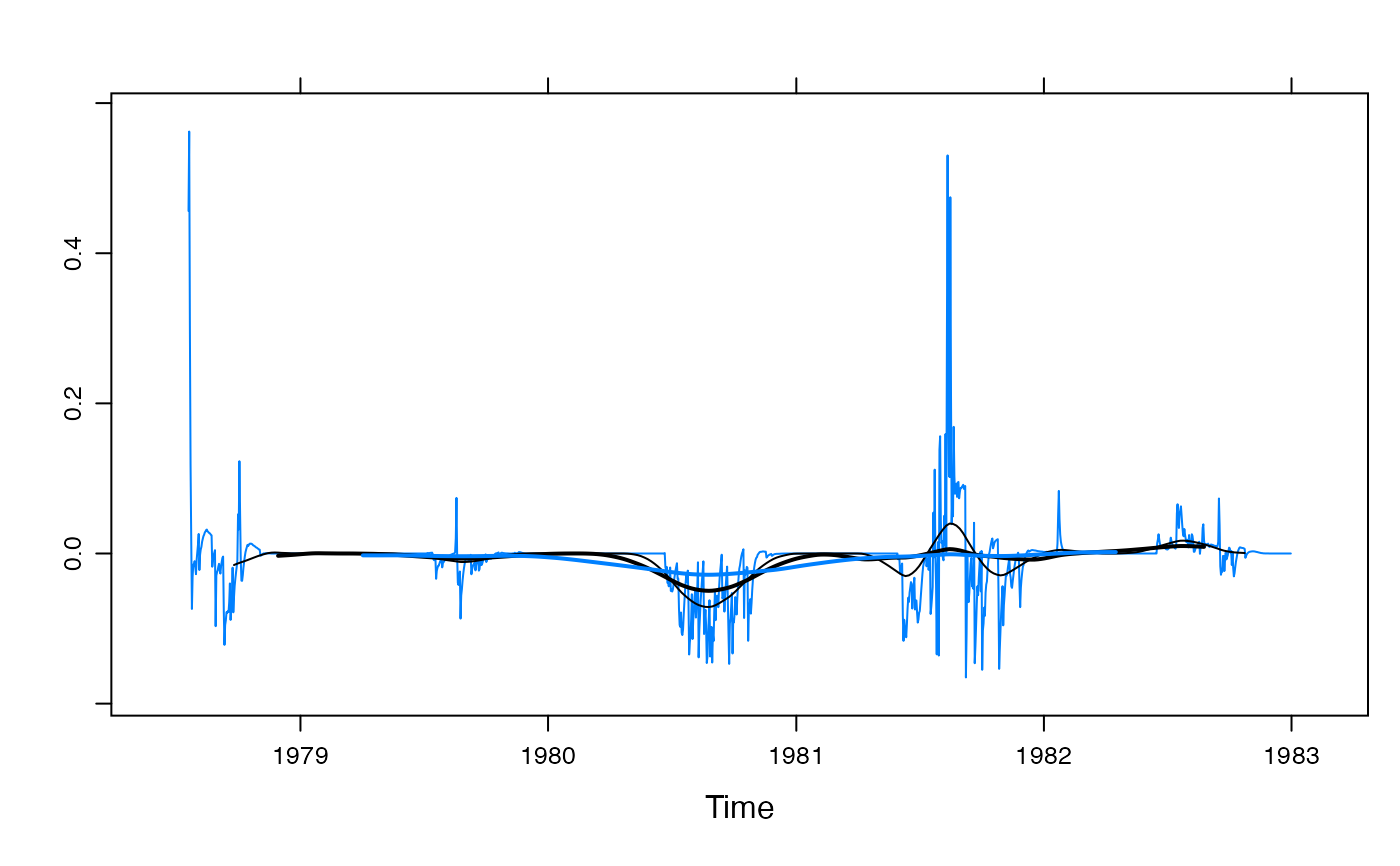
 xyplot(residuals(mod)) +
latticeExtra::layer(panel.tskernel(..., width = 90, c = 2, col = 1)) +
latticeExtra::layer(panel.tskernel(..., width = 180, c = 2, col = 1, lwd = 2)) +
latticeExtra::layer(panel.tskernel(..., width = 360, c = 2, lwd = 2))
xyplot(residuals(mod)) +
latticeExtra::layer(panel.tskernel(..., width = 90, c = 2, col = 1)) +
latticeExtra::layer(panel.tskernel(..., width = 180, c = 2, col = 1, lwd = 2)) +
latticeExtra::layer(panel.tskernel(..., width = 360, c = 2, lwd = 2))
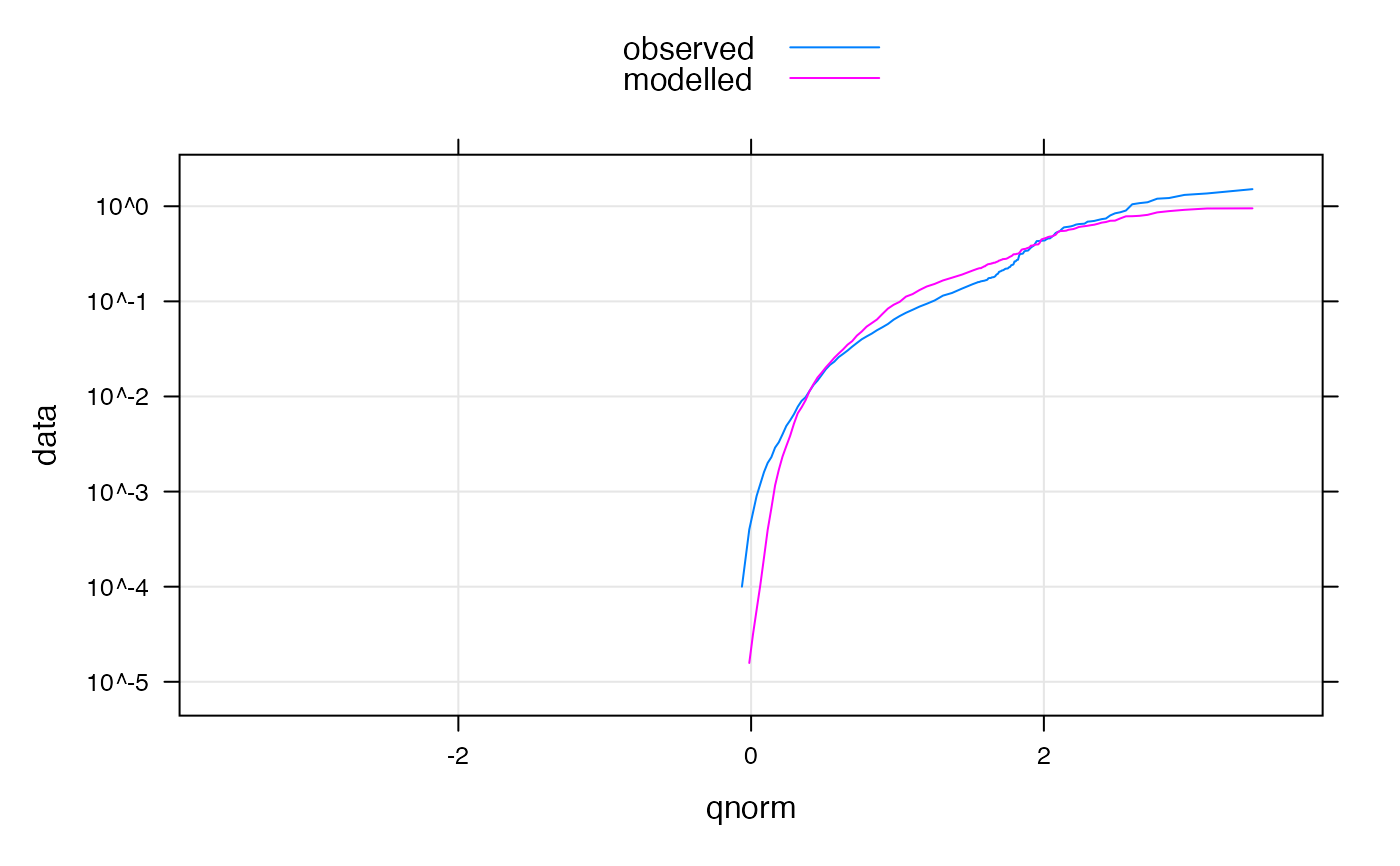
 qqmath(mod,
scales = list(y = list(log = TRUE)), distribution = qnorm,
type = c("g", "l")
)
qqmath(mod,
scales = list(y = list(log = TRUE)), distribution = qnorm,
type = c("g", "l")
)