Support for sensitivity analysis in hydromad
hydromad_sensitivity.RdSensitivity analysis can be performed on hydromad objects using the sensitivity package. The sensitivity analysis functions (e.g. morris, sobol2007) are called directly, specifying the arguments model, object and objective as described below.
Arguments
- model
Must be
model=evalPars, which evaluates theobjectiveusing the model specified byobject- object
an object of class
hydromad.- objective
the objective function or expression, which can refer to Q and X. See
objFunVal.hydromad- parallel
(optional) parallelisation of model runs, see
evalParsandhydromad_parallelisation
References
Shin, Mun-Ju, Joseph H.A. Guillaume, Barry F.W. Croke, and Anthony J. Jakeman. 2013. "Addressing Ten Questions about Conceptual Rainfall-runoff Models with Global Sensitivity Analyses in R." Journal of Hydrology 503 (October): 135-52. doi:10.1016/j.jhydrol.2013.08.047
See also
Utilities evalPars and getFreeParsRanges. tellTS for calculating sensitivity indices on a time series.
Examples
library(sensitivity)
#> Registered S3 method overwritten by 'sensitivity':
#> method from
#> print.src dplyr
#>
#> Attaching package: ‘sensitivity’
#> The following object is masked from ‘package:car’:
#>
#> scatterplot
#> The following object is masked from ‘package:hydromad’:
#>
#> parameterSets
## Load data
data(Cotter)
obs <- Cotter[1:1000]
## Define rainfall-runoff model structure
model.str <- hydromad(obs, sma = "cwi", routing = "expuh",
tau_s = c(2,100), v_s = c(0,1))
## Set the random seed to obtain replicable results
set.seed(19)
################################################################################
## Sensitivity using Morris method of NSE* objective function to
## IHACRES-CWI model parameters using a subset of data from Cotter catchment
## Run Morris Sensitivity analysis
mm <- morris(
## Utility function to evaluate parameters on a model
model=evalPars,
## Names of factors/parameters
factors=names(getFreeParsRanges(model.str)),
## Number of repetitions
r=4,
## List specifying design type and its parameters
design=list(type="oat",levels=10,grid.jump=2),
## Minimum value of each non-fixed parameter
binf=sapply(getFreeParsRanges(model.str),min),
## Maximum value of each non-fixed parameter
bsup=sapply(getFreeParsRanges(model.str),max),
## Hydromad model object to use to evaluate parameters
object=model.str,
## NSE* objective function
objective=~ hmadstat("r.squared")(Q, X) /
(2-hmadstat("r.squared")(Q, X))
)
print(mm)
#>
#> Call:
#> morris(model = evalPars, factors = names(getFreeParsRanges(model.str)), r = 4, design = list(type = "oat", levels = 10, grid.jump = 2), binf = sapply(getFreeParsRanges(model.str), min), bsup = sapply(getFreeParsRanges(model.str), max), object = model.str, objective = ~hmadstat("r.squared")(Q, X)/(2 - hmadstat("r.squared")(Q, X)))
#>
#> Model runs: 20
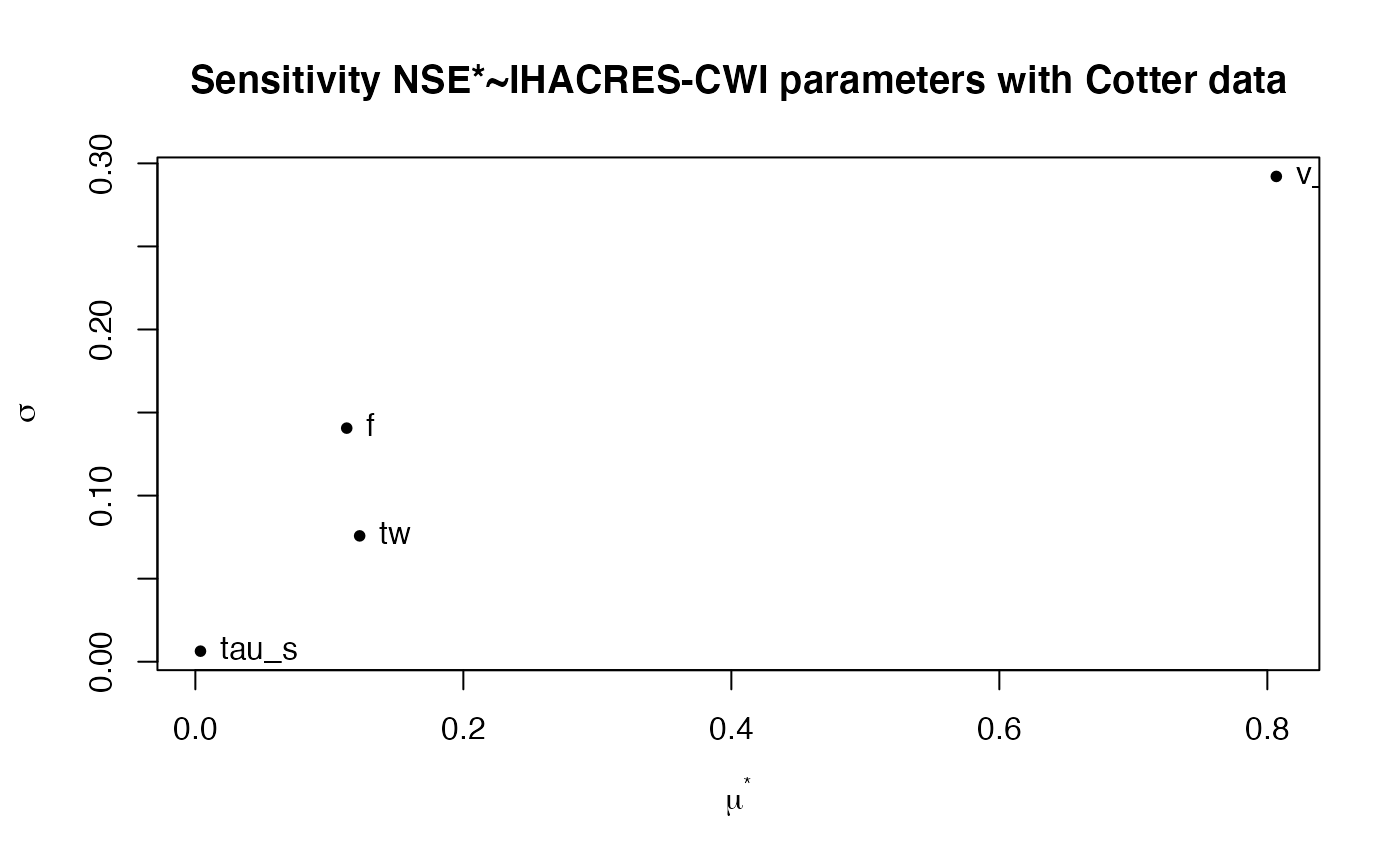
#> mu mu.star sigma
#> tw 0.122615106 0.12261511 0.075759604
#> f -0.018312410 0.11296363 0.140619276
#> tau_s 0.001042739 0.00381977 0.006352805
#> v_s 0.806706980 0.80670698 0.292138811
## Default plot of mu.star (mean absolute sensitivity) and sigma (sd of sensitivity)
plot(mm,main = "Sensitivity NSE*~IHACRES-CWI parameters with Cotter data")
 ## For custom plots, mu.star and sigma can be explicitly calculated
mu.star <- apply(mm$ee, 2, function(x) mean(abs(x)))
sigma <- apply(mm$ee, 2, sd)
plot(mu.star, sigma, pch = 20, xlab = expression(mu^"*"),
ylab = expression(sigma))
text(mu.star, sigma, labels = colnames(mm$ee), pos = 4,offset=0.4)
## For custom plots, mu.star and sigma can be explicitly calculated
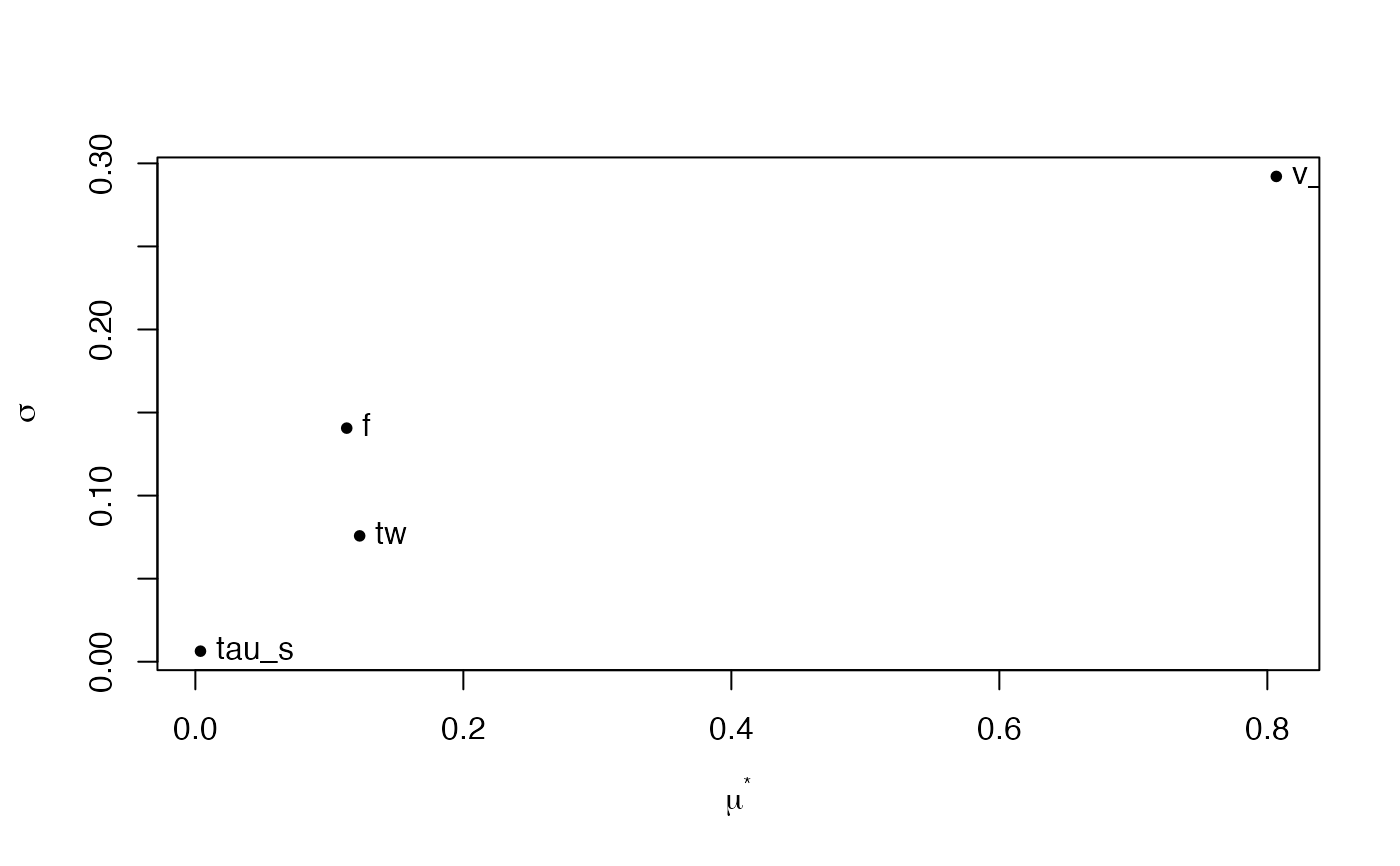
mu.star <- apply(mm$ee, 2, function(x) mean(abs(x)))
sigma <- apply(mm$ee, 2, sd)
plot(mu.star, sigma, pch = 20, xlab = expression(mu^"*"),
ylab = expression(sigma))
text(mu.star, sigma, labels = colnames(mm$ee), pos = 4,offset=0.4)
 ################################################################################
## Sensitivity using SOBOL2002 method of NSElog* prediction function to
## IHACRES-CWI model parameters using a subset of data from Cotter catchment
## This might take a while, potentially ~15min
if (FALSE) {
n <- 1000 ## Number of random samples for parameters
ss <- sobol2002(model = evalPars,
## Draw two random samples of parameters
X1 = parameterSets(getFreeParsRanges(model.str),n),
X2 = parameterSets(getFreeParsRanges(model.str),n),
## Number of bootstrap replicates
nboot = 100,
## Hydromad model object to use to evaluate parameters
object=model.str,
## NSElog* objective function (using the logarithm of Q and X)
objective=~ hmadstat("r.sq.log")(Q, X) /
(2-hmadstat("r.sq.log")(Q, X))
)
## Show results
print(ss)
plot(ss)
}
################################################################################
## Sensitivity using SOBOL2002 method of NSElog* prediction function to
## IHACRES-CWI model parameters using a subset of data from Cotter catchment
## This might take a while, potentially ~15min
if (FALSE) {
n <- 1000 ## Number of random samples for parameters
ss <- sobol2002(model = evalPars,
## Draw two random samples of parameters
X1 = parameterSets(getFreeParsRanges(model.str),n),
X2 = parameterSets(getFreeParsRanges(model.str),n),
## Number of bootstrap replicates
nboot = 100,
## Hydromad model object to use to evaluate parameters
object=model.str,
## NSElog* objective function (using the logarithm of Q and X)
objective=~ hmadstat("r.sq.log")(Q, X) /
(2-hmadstat("r.sq.log")(Q, X))
)
## Show results
print(ss)
plot(ss)
}