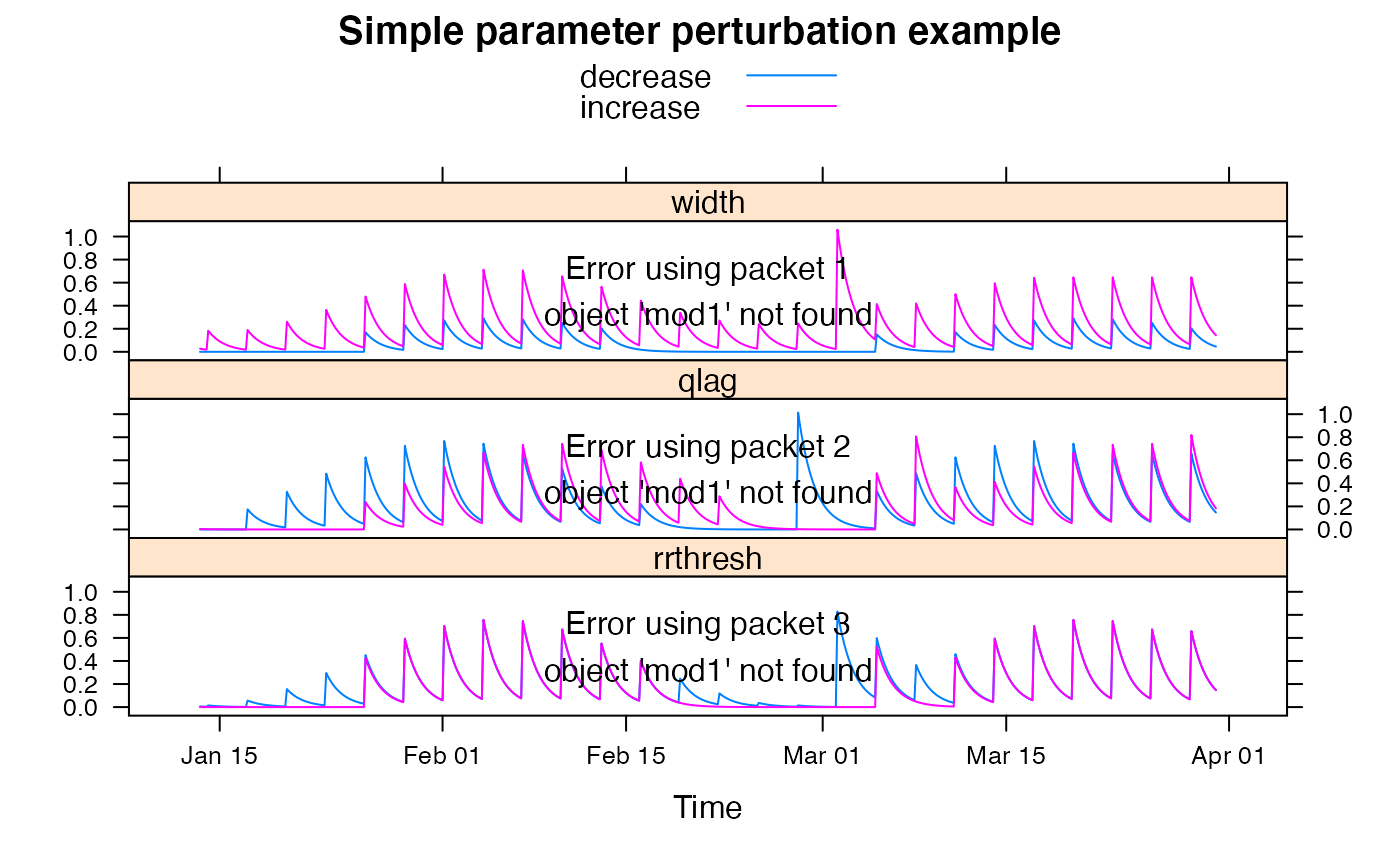
Simple time-varying runoff proportion. Rainfall is scaled by the runoff coefficient estimated in a moving window. This SMA uses streamflow data, so can not be used for prediction.
runoffratio.sim(
DATA,
width = 30,
kernel = 2,
sides = 2,
rrthresh = 0,
qlag = 0,
scale = 1,
return_state = FALSE
)Arguments
- DATA
time-series-like object with columns
P(precipitation) andQ(streamflow).- width
width of the time window (in time steps) in which to estimate the runoff coefficient.
- kernel
type of window used to estimate the runoff coefficient: 1 is rectangular, 2 is triangular-weighted, 3 is Gaussian-like.
- sides
2 for time-centered estimates, 1 for estimates using data backward in time only.
- rrthresh
a theshold value of the runoff ratio, below which there is no effective rainfall.
- qlag
number of time steps to lag the streamflow (relative to rainfall) before estimating the runoff coefficient.
- scale
constant multiplier of the result, for mass balance. If this parameter is set to
NA(as it is by default) inhydromadit will be set by mass balance calculation.- return_state
ignored.
Value
the simulated effective rainfall, a time series of the same length as the input series.
See also
hydromad(sma = "runoffratio") to work with models as
objects (recommended).
Examples
## view default parameter ranges:
str(hydromad.options("runoffratio"))
#> List of 1
#> $ runoffratio:List of 2
#> ..$ rrthresh: num [1:2] 0 0.2
#> ..$ scale : logi NA
data(HydroTestData)
mod0 <- hydromad(HydroTestData, sma = "runoffratio", routing = "expuh")
mod0
#>
#> Hydromad model with "runoffratio" SMA and "expuh" routing:
#> Start = 2000-01-01, End = 2000-03-31
#>
#> SMA Parameters:
#> lower upper
#> rrthresh 0 0.2
#> scale NA NA
#> Routing Parameters:
#> NULL
## simulate with some arbitrary parameter values
mod1 <- update(mod0, width = 30, rrthresh = 0.2, tau_s = 10)
## plot results with state variables
testQ <- predict(mod1, return_state = TRUE)
xyplot(cbind(HydroTestData[, 1:2], runoffratio = testQ))
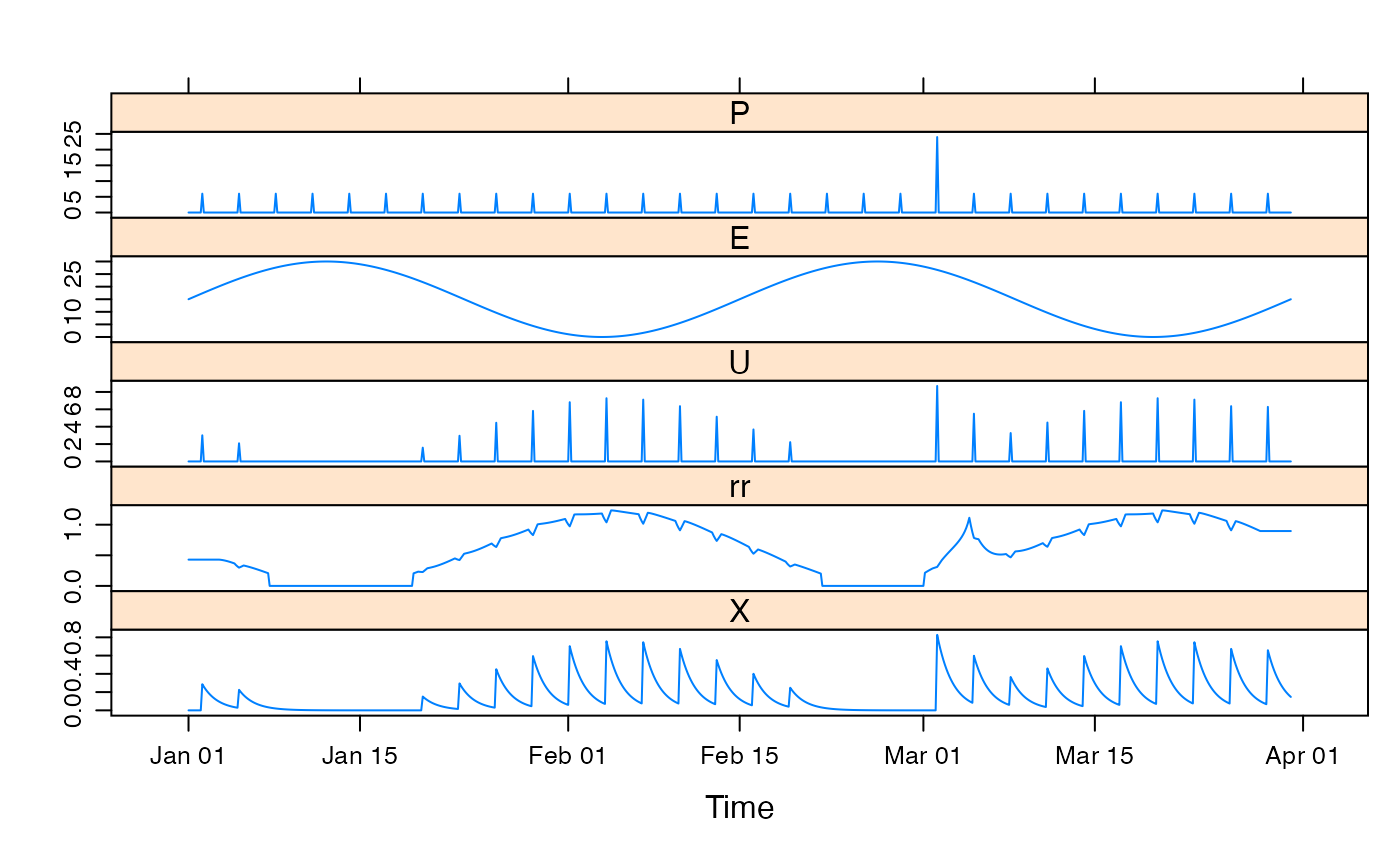 ## show effect of increase/decrease in each parameter
parRanges <- list(
width = c(10, 180), qlag = c(-30, 30),
rrthresh = c(0, 0.5)
)
parsims <- mapply(
val = parRanges, nm = names(parRanges),
FUN = function(val, nm) {
lopar <- min(val)
hipar <- max(val)
names(lopar) <- names(hipar) <- nm
fitted(runlist(
decrease = update(mod1, newpars = lopar),
increase = update(mod1, newpars = hipar)
))
}, SIMPLIFY = FALSE
)
xyplot.list(parsims,
superpose = TRUE, layout = c(1, NA),
main = "Simple parameter perturbation example"
) +
latticeExtra::layer(panel.lines(fitted(mod1), col = "grey", lwd = 2))
## show effect of increase/decrease in each parameter
parRanges <- list(
width = c(10, 180), qlag = c(-30, 30),
rrthresh = c(0, 0.5)
)
parsims <- mapply(
val = parRanges, nm = names(parRanges),
FUN = function(val, nm) {
lopar <- min(val)
hipar <- max(val)
names(lopar) <- names(hipar) <- nm
fitted(runlist(
decrease = update(mod1, newpars = lopar),
increase = update(mod1, newpars = hipar)
))
}, SIMPLIFY = FALSE
)
xyplot.list(parsims,
superpose = TRUE, layout = c(1, NA),
main = "Simple parameter perturbation example"
) +
latticeExtra::layer(panel.lines(fitted(mod1), col = "grey", lwd = 2))