Fit a hydromad model using the DE (Differential Evolution) algorithm.
Source:R/fitByDE.R
fitByDE.RdFit a hydromad model using the DE (Differential Evolution) algorithm.
fitByDE(
MODEL,
objective = hydromad.getOption("objective"),
control = hydromad.getOption("de.control")
)Arguments
- MODEL
a model specification created by
hydromad. It should not be fully specified, i.e one or more parameters should be defined by ranges of values rather than exact values.- objective
objective function to maximise, given as a
function(Q, X, ...). SeeobjFunVal.- control
settings for the DE algorithm. See
DEoptim.control.
Value
the best model from those sampled, according to the given
objective function. Also, these extra elements are inserted:
- fit.result
the result from
DEoptim.- objective
the
objectivefunction used.- funevals
total number of evaluations of the model simulation function.
- timing
timing vector as returned by
system.time.
Examples
library("DEoptim")
#> Loading required package: parallel
#>
#> DEoptim package
#> Differential Evolution algorithm in R
#> Authors: D. Ardia, K. Mullen, B. Peterson and J. Ulrich
data(Cotter)
x <- Cotter[1:1000]
## IHACRES CWI model with power law unit hydrograph
modx <- hydromad(x, sma = "cwi", routing = "powuh")
modx
#>
#> Hydromad model with "cwi" SMA and "powuh" routing:
#> Start = 1966-05-01, End = 1969-01-24
#>
#> SMA Parameters:
#> lower upper
#> tw 0 100
#> f 0 8
#> scale NA NA
#> l 0 0 (==)
#> p 1 1 (==)
#> t_ref 20 20 (==)
#> Routing Parameters:
#> lower upper
#> a 0.01 60
#> b 0.50 3
#> c 0.50 2
foo <- fitByDE(modx, control = DEoptim.control(itermax = 5))
summary(foo)
#>
#> Call:
#> hydromad(DATA = x, sma = "cwi", routing = "powuh", a = 8.4263,
#> b = 1.25033, c = 1.76043, tw = 96.2573, f = 5.15738, scale = 0.00131429)
#>
#> Time steps: 900 (0 missing).
#> Runoff ratio (Q/P): (0.7028 / 2.285) = 0.3075
#> rel bias: -3.69e-18
#> r squared: 0.7309
#> r sq sqrt: 0.8295
#> r sq log: 0.8256
#>
#> For definitions see ?hydromad.stats
#>
## return value from DE:
str(foo$fit.result)
#> List of 2
#> $ optim :List of 4
#> ..$ bestmem: Named num [1:5] 8.43 1.25 1.76 96.26 5.16
#> .. ..- attr(*, "names")= chr [1:5] "a" "b" "c" "tw" ...
#> ..$ bestval: num -0.851
#> ..$ nfeval : int 12
#> ..$ iter : int 5
#> $ member:List of 6
#> ..$ lower : Named num [1:5] 0.01 0.5 0.5 0 0
#> .. ..- attr(*, "names")= chr [1:5] "a" "b" "c" "tw" ...
#> ..$ upper : Named num [1:5] 60 3 2 100 8
#> .. ..- attr(*, "names")= chr [1:5] "a" "b" "c" "tw" ...
#> ..$ bestmemit: num [1:5, 1:5] 10.99 10.99 10.99 10.99 8.43 ...
#> .. ..- attr(*, "dimnames")=List of 2
#> .. .. ..$ : chr [1:5] "1" "2" "3" "4" ...
#> .. .. ..$ : chr [1:5] "a" "b" "c" "tw" ...
#> ..$ bestvalit: num [1:5] -0.842 -0.842 -0.847 -0.847 -0.851
#> ..$ pop : num [1:50, 1:5] 12.4 25.7 10.5 24.9 13.2 ...
#> ..$ storepop : list()
#> - attr(*, "class")= chr "DEoptim"
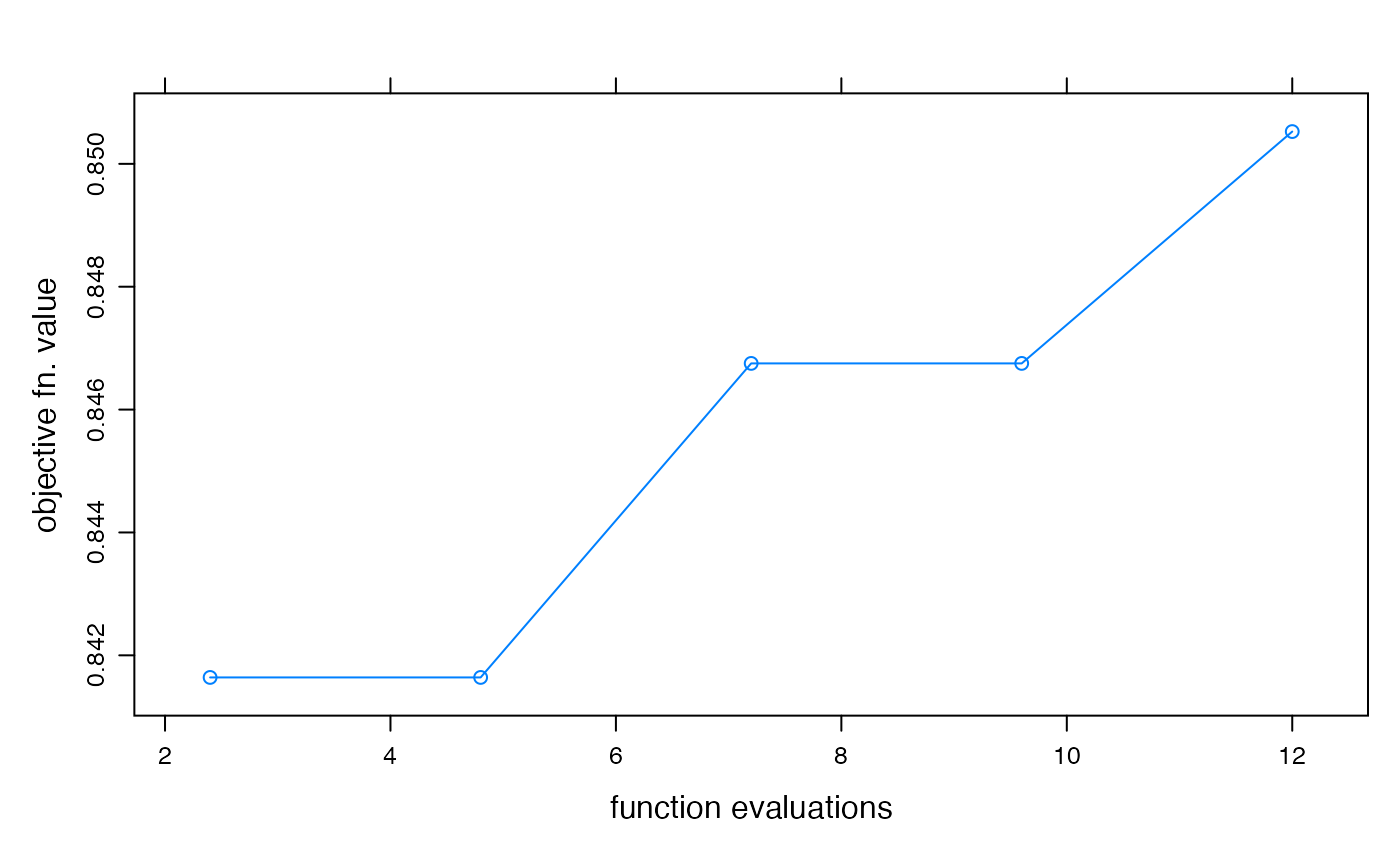
## plot objective function value convergence over time
xyplot(optimtrace(foo),
type = "b",
xlab = "function evaluations", ylab = "objective fn. value"
)