The Catchment Moisture Deficit (CMD) effective rainfall model for IHACRES. It is a conceptual-type model, where input rainfall is partitioned explicitly into drainage, evapo-transpiration, and changes in catchment moisture.
cmd.sim(DATA, f, e, d, shape = 0, M_0 = d/2, return_state = FALSE)Arguments
- DATA
a
ts-like object with named columns:- list("P")
time series of areal rainfall depths, usually in mm.
- list("E")
time series of potential evapo-transpiration, or more typically, temperature as an indicator of this.
- f
CMD stress threshold as a proportion of
d.- e
temperature to PET conversion factor.
- d
CMD threshold for producing flow.
- shape
defines form of the \(dU/dP\) relationship:
shape = 0is the linear form,shape = 1is the trigonometric form, andshape > 1is the power form.- M_0
starting CMD value.
- return_state
to return state variables as well as the effective rainfall.
Value
cmd.sim returns the modelled time series of effective
rainfall, or if return_state = TRUE, a multi-variate time series with
named columns U (effective rainfall), CMD and ET(evapo-transpiration \(E_T\)).
Details
The mass balance step is: $$M[t] = M[t-1] - P[t] + E_T[t] + U[t]$$
where \(M\) represents catchment moisture deficit (CMD), constrained below by 0 (the nominal fully saturated level). P is catchment areal rainfall, \(E_T\) is evapo-transpiration, and U is drainage (effective rainfall). All are, typically, in units of mm per time step.
Rainfall effectiveness (i.e. drainage proportion) is a simple instantaneous function of the CMD, with a threshold at \(M = d\). In the default linear form this is:
$$\frac{\mathrm{d}U}{\mathrm{d}P} = 1 - \min(1, M/d)$$
The trigonometric form is
$$\frac{\mathrm{d}U}{\mathrm{d}P} = 1 - \min(1, \sin^2(\pi M / 2d))$$
The power form is
$$\frac{\mathrm{d}U}{\mathrm{d}P} = 1 - \min(1, (M/d)^a)$$ where a = 10 ^ (shape / 50)
The actual drainage each time step involves the integral of these relations.
Evapo-transpiration is also a simple function of the CMD, with a threshold at \(M = f d\): $$E_T[t] = e E[t] \min(1, \exp\left(2\left(1 - \frac{M_f}{fd}\right)\right))$$
Note that the evapo-transpiration calculation is based on \(M_f\), which is the CMD after precipitation and drainage have been accounted for.
Note
Normally compiled C code is used for simulation, but if
return_state = TRUE a slower implementation in R is used.
References
Croke, B.F.W. and A.J. Jakeman (2004), A Catchment Moisture Deficit module for the IHACRES rainfall-runoff model, Environmental Modelling and Software, 19(1): 1-5.
Croke, B.F.W. and A.J. Jakeman (2005), Corrigendum to ``A Catchment Moisture Deficit module for the IHACRES rainfall-runoff model'' [Environ. Model. Softw. 19 (1) (2004) 1-5], Environmental Modelling and Software, 20(7): 977.
See also
hydromad(sma = "cmd") to work with models as objects
(recommended).
Examples
## view default parameter ranges:
str(hydromad.options("cmd"))
#> List of 1
#> $ cmd:List of 4
#> ..$ f : num [1:2] 0.01 3
#> ..$ e : num [1:2] 0.01 1.5
#> ..$ d : num [1:2] 50 550
#> ..$ shape: num 0
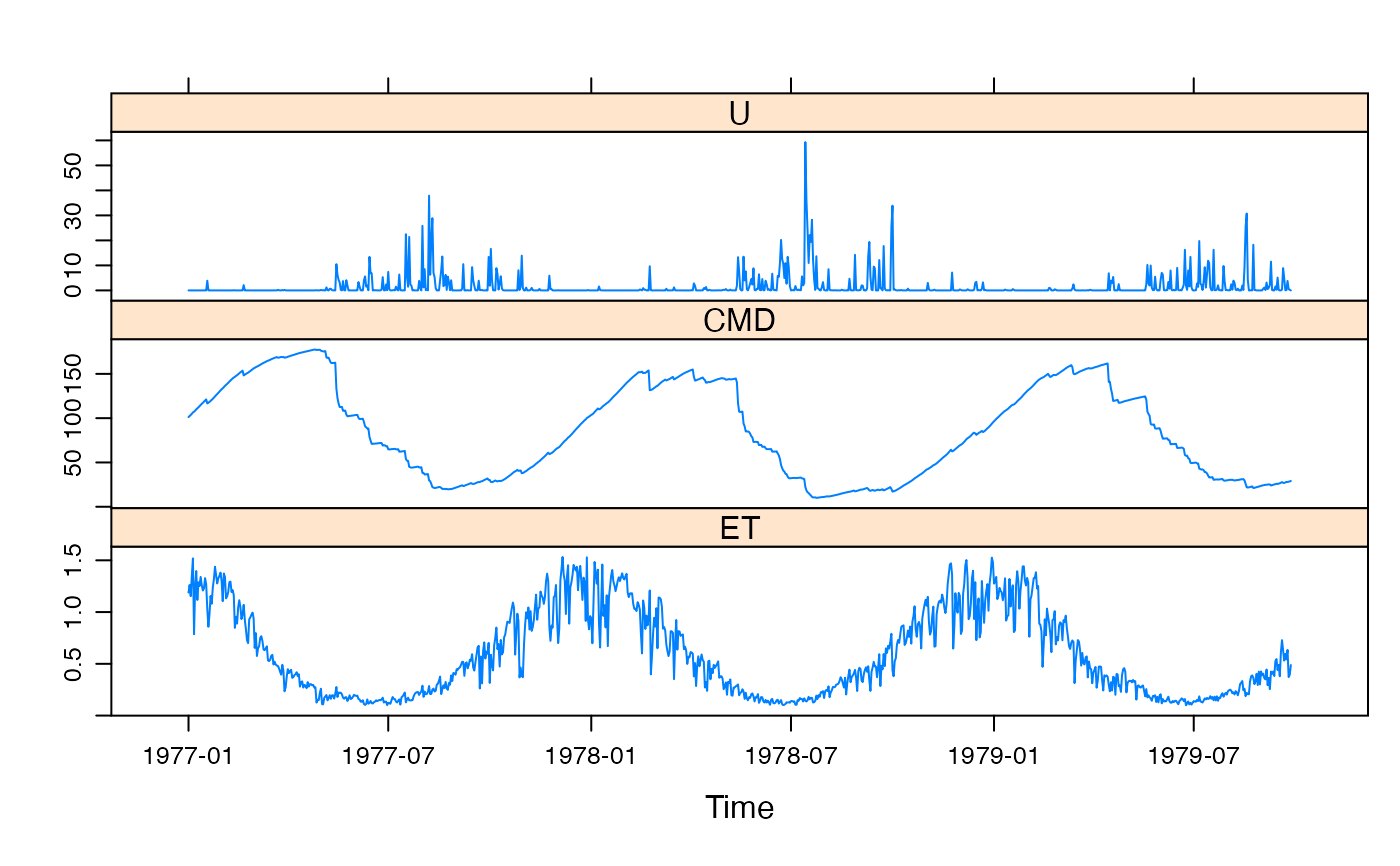
data(Canning)
x <- cmd.sim(Canning[1:1000, ],
d = 200, f = 0.7, e = 0.166,
return_state = TRUE
)
xyplot(x)
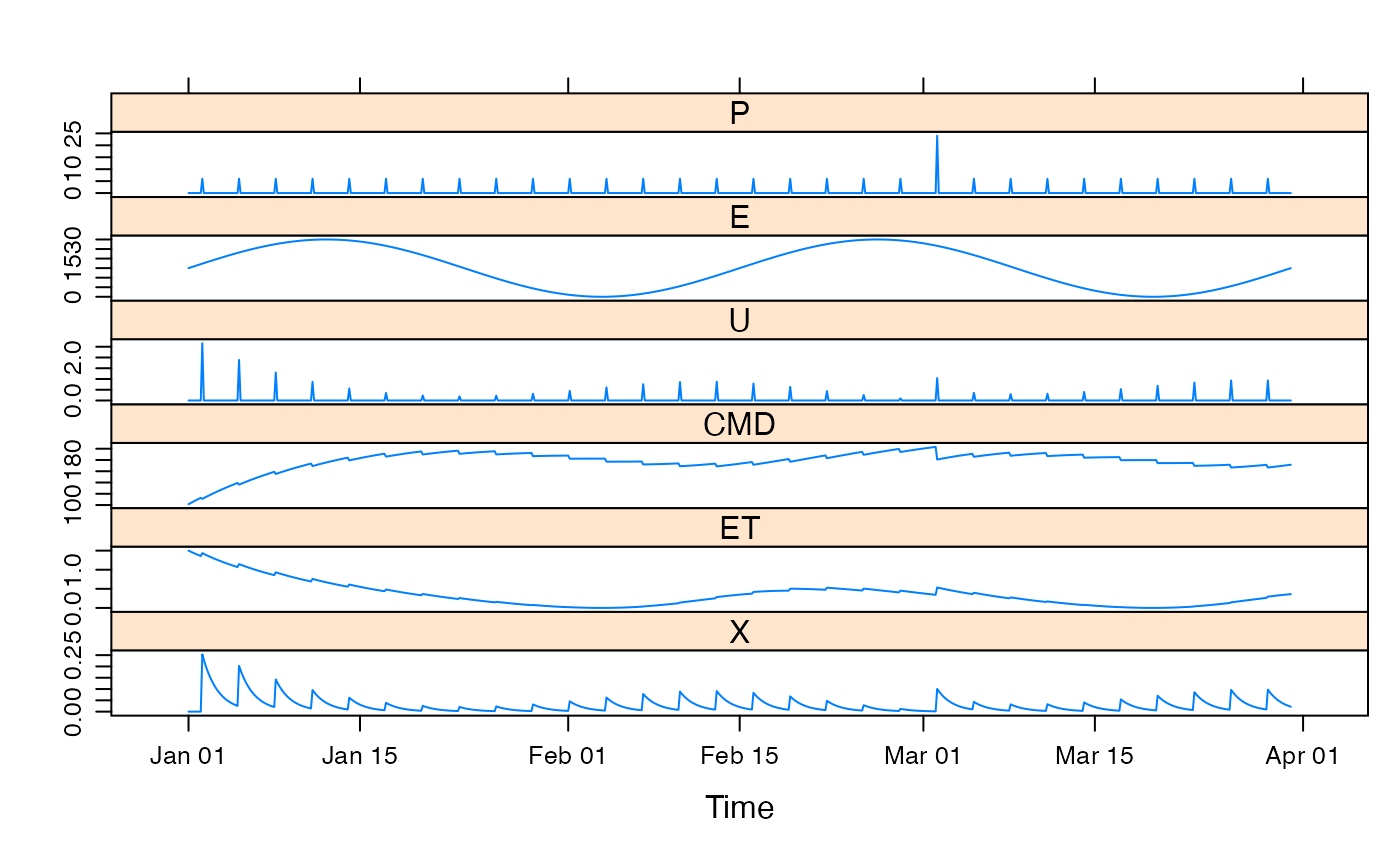
 data(HydroTestData)
mod0 <- hydromad(HydroTestData, sma = "cmd", routing = "expuh")
mod0
#>
#> Hydromad model with "cmd" SMA and "expuh" routing:
#> Start = 2000-01-01, End = 2000-03-31
#>
#> SMA Parameters:
#> lower upper
#> f 0.01 3.0
#> e 0.01 1.5
#> d 50.00 550.0
#> shape 0.00 0.0 (==)
#> Routing Parameters:
#> NULL
## simulate with some arbitrary parameter values
mod1 <- update(mod0, d = 200, f = 0.5, e = 0.1, tau_s = 10)
## plot results with state variables
testQ <- predict(mod1, return_state = TRUE)
xyplot(cbind(HydroTestData[, 1:2], cmd = testQ))
data(HydroTestData)
mod0 <- hydromad(HydroTestData, sma = "cmd", routing = "expuh")
mod0
#>
#> Hydromad model with "cmd" SMA and "expuh" routing:
#> Start = 2000-01-01, End = 2000-03-31
#>
#> SMA Parameters:
#> lower upper
#> f 0.01 3.0
#> e 0.01 1.5
#> d 50.00 550.0
#> shape 0.00 0.0 (==)
#> Routing Parameters:
#> NULL
## simulate with some arbitrary parameter values
mod1 <- update(mod0, d = 200, f = 0.5, e = 0.1, tau_s = 10)
## plot results with state variables
testQ <- predict(mod1, return_state = TRUE)
xyplot(cbind(HydroTestData[, 1:2], cmd = testQ))
 ## show effect of increase/decrease in each parameter
parlist <- list(
d = c(50, 550), f = c(0.01, 3),
e = c(0.01, 1.5)
)
parsims <- mapply(
val = parlist, nm = names(parlist),
FUN = function(val, nm) {
lopar <- min(val)
hipar <- max(val)
names(lopar) <- names(hipar) <- nm
fitted(runlist(
decrease = update(mod1, newpars = lopar),
increase = update(mod1, newpars = hipar)
))
}, SIMPLIFY = FALSE
)
xyplot.list(parsims,
superpose = TRUE, layout = c(1, NA),
main = "Simple parameter perturbation example"
) +
latticeExtra::layer(panel.lines(fitted(mod1), col = "grey", lwd = 2))
## show effect of increase/decrease in each parameter
parlist <- list(
d = c(50, 550), f = c(0.01, 3),
e = c(0.01, 1.5)
)
parsims <- mapply(
val = parlist, nm = names(parlist),
FUN = function(val, nm) {
lopar <- min(val)
hipar <- max(val)
names(lopar) <- names(hipar) <- nm
fitted(runlist(
decrease = update(mod1, newpars = lopar),
increase = update(mod1, newpars = hipar)
))
}, SIMPLIFY = FALSE
)
xyplot.list(parsims,
superpose = TRUE, layout = c(1, NA),
main = "Simple parameter perturbation example"
) +
latticeExtra::layer(panel.lines(fitted(mod1), col = "grey", lwd = 2))